

To elucidate the structure of N-linked oligosaccharides, we previously proposed a three-dimensional (3-D) sugar mapping technique using pyridylaminated N-linked neutral and sialyl glycans. This is a powerful separation technique and can estimate the oligosaccharide structures on the basis of the elution positions on threee HPLC columns using only pico-moles of samples. The elution coordinate database of this web site is necessary for the 2-D/3-D sugar mapping techniques. It contains more than 400 pyridylaminated N-glycan structures, code numbers, the elution positions on Shimpack CLC-ODS and Amide-80 columns, sources and references. All the data presented here have been obtained by our group.
Below is an example of the database content.
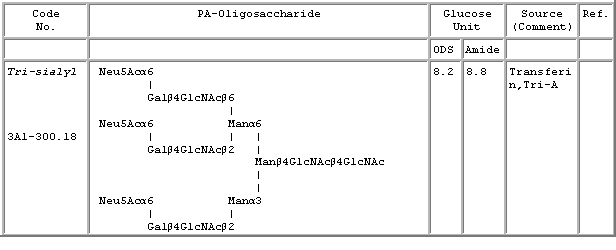
Elution position: The elution positions of the PA-glycans in this database are expressed in the glucose units, in reference to the PA derivatized isomalto-oligosaccharides of DP 3-25 (Tomiya, N., (1988) Anal. Biochem., 171, 73-90.).
Code number: The code number of the PA-N-glycans consists of a set of several elements with the following meanings. The number to the right of the hyphen (here, 310.8) represents the neutral N-glycan code number (ref. Tomiya, N. et al., (1988) Anal Biochem. 171, 73-90). Asterisk (*) indicate that the structure was confirmed by NMR.

(1) Neutral complex type PA-glycans 1) Core and mono-antennary N-glycans 2) Bi-antennary N-glycans 3) Tri-antennary N-glycans 4) Tetra- and penta-antennary N-glycans 5) GalNAc-containing N-glycans (2) Sialyl complex type PA-glycans with Neu5Ac (A) Mono-sialyl N-glycans 1) Mono-antennary N-glycans 2) Bi-antennary N-glycans 3) Tri-antennary N-glycans 4) Tetra-antennary N-glycans (B) Di-sialyl N-glycans 1) Bi-antennary N-glycans 2) Tri-antennary N-glycans 3) Tetra-antennary N-glycans (C) Tri-sialyl N-glycans 1) Tri-antennary N-glycans 2) Tetra-antennary N-glycans (D) Tetra-sialyl N-glycans 1) Tri-antennary N-glycans 2) Tetra-antennary N-glycans (3) Sialyl complex-type PA-glycans with Neu5Gc (4) High-mannose type PA-glycans (5) Hybrid type PA-glycans (6) Other PA-glycans from plants and insects
The ECD files can be obtained by downloading one of the following files. For WordPerfect users; *WordPerfect (6/7/8 ) file (373KB) *Zip compressed file (42KB) For Microsoft Word users; *Microsoft Word (6/7) file (394KB) *Zip compressed file (70KB) *Lha compressed file (67KB)
1. Separate PA-glycan mixture on a DEAE column (1st column) according to its sialic acid content, into neutral, mono-, di-, tri-sialyl N-glycans, etc., and evaporate separately.
2. Calibrate both the ODS (2nd column) and amide (3rd column) columns with PA-derivatized isomalto-oligosaccharide mixtures (standard glucose oligomers). Numbers 4, 5, 6, etc., indicate the degree of polymerization of glucose.
3. Apply each sample PA-glycan separated on the DEAE column, onto the calibrated ODS column, and express the elution volume as a glucose unit to be plotted on the X-axis.
4. Apply each sample PA-glycan separated on the ODS column, onto the Amide-80 column, and express the elution volumn as a glucose unit to be plotted on the Y-axis.
5. Plot the coordinate of the sample PA-glycan on the 2-D map.
6. Repeat above processes 3-5, for each group of different sialylation and form a 3-D map (Takahashi, N. et al., (1995) Anal. Biochem. 226, 139-146.; Takahashi N (1996) J. Chromatogr. A. 720, 217-225.).
The all HPLC elution conditions used to obtain the X-, and Y-coordinates for sialyl glycans are completely the same as described previously for neutral glycans (Tomiya, N. et al., (1988) Anal. Biochem., 171, 73-90.).
The structure of an unknown PA-glycan is estimated by comparing its position on the map with the positions of the known reference PA-glycans.
1. Choose a few candidate PA-glycans whose coordinates coincide with that of the smple PA-glycan within allowable error (5%), out of the database, by using a computer search.
2. Co-inject the sample PA-glycan and one of the candidate standard PA-glycans (whose structure has been already established) into two different HPLC columns and confirm to give a single symmetry peak.
3. Digest the sample PA-glycan and the candidate standard PA-glycan with several glycosidases simultaneously. Compare again their elution positions. Continue the comparison until both PA-glycans yiel the common trimannosyl core (Takahashi, N. and Tomiya, N. (1992) Handbook of Endoglycosidases and Glycoamidases, p. 199-332.; Tomiya, N. et al., (1991) Anal. Biochem., 193, 90-100.; Nakagawa, H. et al., (1996) Eur. J. Biochem., 237, 76-85.).